Using machine learning and image data to study bacterial communities
The word data is thrown around a lot these days. But what counts as data? Numbers? Words?
What about images?
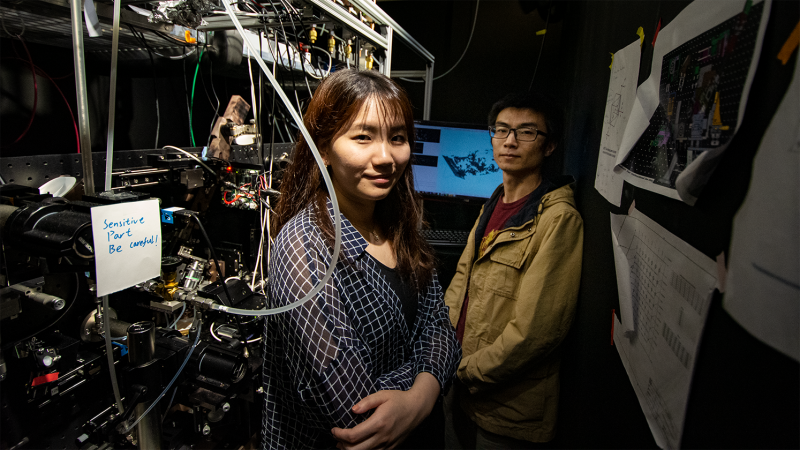
University of Virginia doctoral students Jie Wang and Mingxing Zhang are using images as a form of data as they work to create and process visualizations of single cells at unprecedented levels of detail.
Specifically, Wang and Zhang are studying bacterial communities, the growth and function of which are made up of aggregates of single cells, and which are central to biomedical and biochemical research. Wang and Zhang hope to improve the way these bacterial communities are studied through a new biofilm method.
This project is one of six projects within the Presidential Fellowships in Data Science program. This fellowship provides opportunities and funding for graduate students across a variety of fields of study to work together on projects that address real-world problems using traditional research methods alongside cutting-edge data science tools and techniques.
Zhang, a Ph.D. candidate in Chemistry, plans to study single bacteria in biofilms using lattice-light sheet microscopy, an advanced and minimally invasive microscopy method. Wang, a Ph.D. candidate in Electrical and Computer Engineering, will then turn the acquired 3-D fluorescence images into quantifiable information, and ultimately, into biological insight.
What is Biofilm?
“Biofilm is a 3-D tissue structure [that adheres to a surface],” Zhang explained. “Biofilm bacteria is a major component of the actual biomass of the Earth. It plays a critical role in the biochemistry of the planet and also the biochemistry of the high-level living organisms, like humans.”
Zhang went on to explain that biofilm is critical in both human health and the environment. He added that bacterial biofilms are estimated to be in 80% of all human bacterial infections. An example of such an infection is cystic fibrosis, a serious disease that can damage lung and digestive health. In the environment, bacterial biofilms play a big role in driving biogeochemical cycles.
Biofilms, however, are difficult to study through conventional methods.
How Biofilm is Studied
Zhang explained that conventional microscopes can exhibit phototoxic effects in light-sensitive living specimens. In other words, using conventional microscopes to study biofilms would be invasive and could impact the growth of the bacteria in the biofilm. Therefore, the resulting images would not display the natural state of the bacterial biofilm.
So is it possible to study the natural state and growth of biofilms in a non-invasive way?
Zhang is using lattice light-sheet microscopy (LLSM) to solve this issue. This process “resolves individual cells within thick 3-D biofilms in a non-invasive manner.” Zhang explained that the illumination intensity of LLSM is comparable to the intensity of the sun, under which life has evolved. LLSM solves this issue of phototoxicity, as it does not alter the growth of biofilm bacteria, while allowing a close-up look at what is happening in the biofilms.
The high spatial and temporal resolution of LLSM allows Zhang to resolve the single cell behaviors within the biofilm. This behavior includes how the cells interact with each other, how they eat, how they move, and how they communicate with each other to coordinate their behaviors.
Zhang is currently studying a bacterial strain called Myxococcus xanthus. Zhang explained that this is a good strain to study for several reasons. One of these reasons is that this soil-dwelling bacterium displays significant social behaviors on a multicellular level. Additionally, M. xanthus can form large fruiting bodies, changing from rod-shaped cells to spherical spores that are highly resistant to environmental stressors.
Not much is known about the actual process of the shape change of M. xanthus. By using LLSM and partnering with Wang, Zhang plans to uncover more about the biochemical signals that initiate this process.
How the Computer Engineering Process Comes In
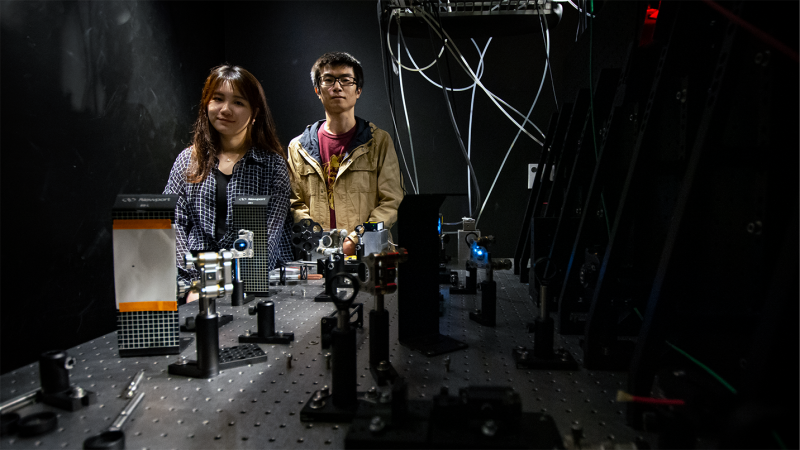
While Zhang uses LLSM to study details of M. xanthus and create images of the bacterial biofilm, Wang processes the images she receives from Zhang using computational engineering.
Wang explained that extracting useful from LLSM images is an exciting challenge for her.
The bacteria undergo a distinctive morphological change when they adopt their spherical shape. Wang is using her computer and electrical engineering skills to visualize this process and to extract useful quantitative information from cell segmentation results, including degree of cell aggregation, change, and growth.
However, Wang’s goal for this project goes beyond processing visualizations for just one type of bacteria. She is working with Zhang to use machine learning to create a more efficient toolkit for this kind of image analysis.
“We want to learn what machine learning can do according to our data set, because we have a lot of data,” Wang explained. “The benefit of machine learning is that once we can apply the method on our data sets, we can apply it later to test on new data sets, which have the same kind of image.”
Wang further explained that 3-D machine learning is not yet perfect, and this image-producing process, which uses Artificial Intelligence (AI), also requires some careful human supervision. Despite the challenges the project presents, Wang and Zhang believe that they are close to creating a 3-D bacterial cell segmentation and image analysis toolkit that will be applicable to a wide range of research.
Teamwork Makes the Dream Work
The beauty behind the Presidential Fellowship Project is that it brings together researchers across different disciplines to approach such challenging projects. Zhang and Wang, with years of experience and expertise in their individual areas of study, are able to assist each other in better understanding the project and their roles in the research.
Zhang explains that he needed Wang to fully understand what he was seeing through the microscope.
“In our field, we just focus on doing the experiment, but we sometimes lack the knowledge to process the data,” Zhang stated.
Wang, on the other hand, needed to understand what the process was behind the images she received in order to process the information the most effectively.
“For us, we get these images, but we normally don’t know what the stages are behind this image,” Wang explained. “So through this collaboration, we go to their [Zhang and his team] lab and see how these images are made and what equipment they use for imaging the data, which is really cool.”
The potential breakthroughs from this project are exciting. A core basis of biomedical, biotechnological, and biochemical research is individual cell behaviors. Cell behaviors and the coordination of such actions and behaviors affect how bacterial communities amass and operate as a population.
Wang and Zhang’s work is poised to have lasting impacts both in biochemical research and computer engineering imaging processes.